Seoul National University Hospital discovers hearing loss mutations and creates a genetic map of hearing loss in Korean
- Integrated genetic analysis, including whole genome sequencing (WGS), improves diagnostic rate by 20% compared to previous tests
- Analysis of non-coding and structural variants and genotype-phenotype associations suggests potential for gene therapy development
The research team at Seoul National University Hospital announced that through an integrated genetic analysis method including whole genome sequencing (WGS), they identified the genetic causes of sensorineural hearing loss (SNHL) and newly constructed a genetic map of hearing loss in Koreans. By utilizing WGS, they achieved a diagnostic rate approximately 20% higher than previous precise analysis methods and made significant new discoveries about the genetic causes of hearing loss. The study results were published in the latest issue of Cell Reports Medicine, an international academic journal specializing in medical research and experimentation.
Hearing loss is classified into conductive hearing loss (caused by problems in the outer and middle ear) and sensorineural hearing loss (caused by nerve transmission problems between the auditory nerve and the brain). Sensorineural hearing loss can result from various causes, including genetic factors, congenital infections, trauma, drug toxicity, and autoimmune diseases. Until now, the genetic causes of hearing loss have been very diverse and complex, making it difficult in many cases to accurately identify the genetic cause. Moreover, previous targeted panel tests and whole-exome sequencing were unable to identify genetic causes in about 50% of patients. This study addressed these limitations by adopting a stepwise approach, incorporating WGS to capture a wider spectrum of genetic variants.
The research team, consisting of Professor Lee Sang-Yeon (Department of Otorhinolaryngology, SNUH), Professors Chae Jong-Hee and Lee Seung-Bok (Department of Clinical Genomic Medicine, SNUH), Dr. Koh June-Young (Inocras), and Dr. Paek Seong-Yeol (Stanford University Genomic Research Laboratory), analyzed 394 families (752 individuals) with hearing loss treated at SNUH’s Department of Otorhinolaryngology and Cochlear Implant Center.
The research team employed a stepwise genetic testing approach. They first identified major genes such as GJB2 using single-gene PCR testing. Then, they analyzed a broader range of genes using targeted panel sequencing (TPS) and whole exome sequencing (WES). In the final stage, whole genome sequencing (WGS) was used to identify structural variants and deep intronic variants (non-coding region variants) that were not detected by previous tests (TPS and WES). This comprehensive approach enabled the discovery of additional variants missed by earlier methods, improving the genetic diagnosis of sensorineural hearing loss.
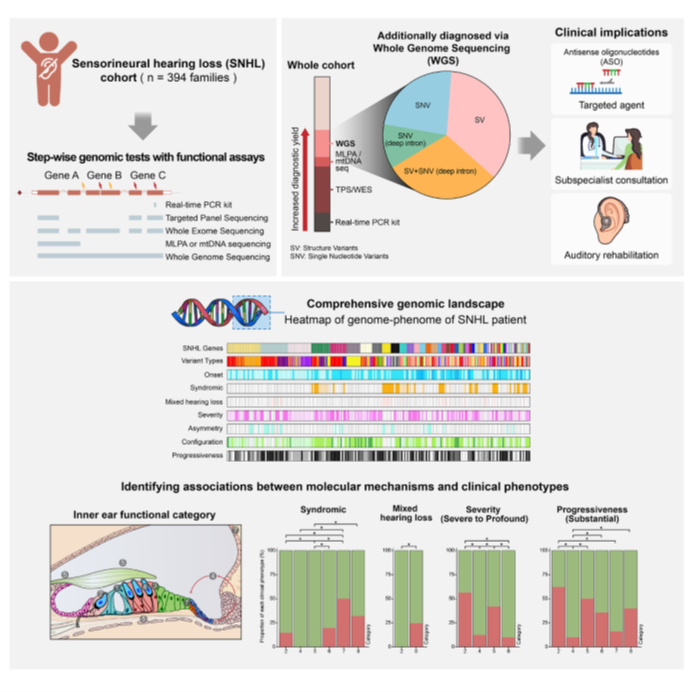
[Figure] Summary of the sensorineural hearing loss genetic analysis process: Constructing a genetic map of hearing loss in Koreans through whole genome sequencing and analyzing the relationships between non-coding and structural variants and types of hearing loss.
As a result of the research, the stepwise approach, including whole genome sequencing, identified the genetic causes in 219 out of 394 families with sensorineural hearing loss. Notably, they discovered an additional 19.2% (44 families) of variants that were not detectable by previous precise tests (targeted panel sequencing, whole exome sequencing), improving the diagnostic rate of hereditary hearing loss by about 20%. This demonstrates that WGS can uncover previously missed variants, substantially enhancing overall diagnostic accuracy.
Additionally, this study was the first to identify non-coding region variants such as deep intronic variants and structural variants. Deep intronic pathogenic variants occur in non-coding regions beyond the boundaries of exons and introns, which play important roles in protein production within genes. These regions could not be detected by previous testing methods. This discovery provided new insights into the genetic causes of hearing loss. In particular, three new deep intronic variants found in the USH2A gene, a representative gene of Usher syndrome, caused splicing errors affecting protein production. Most importantly, this breakthrough highlights the potential for RNA-based gene therapies targeting deep intronic variants.
By providing a genetic map of hearing loss in Koreans, the study demonstrates that WGS can be a critical tool for accurately identifying genetic causes of hearing loss. The team offered a detailed variant map and conducted genotype-phenotype analyses that deepen the understanding of the diverse clinical features of hearing loss. This research paves the way for developing gene-based personalized therapies.
Professor Lee Sang-Yeon (Department of Otorhinolaryngology, SNUH) said, “Through this study, we were able to newly identify the causes of many undiagnosed hearing loss patients and discover patient groups eligible for gene therapy. Going forward, we will actively use WGS to resolve undiagnosed causes of hearing loss and establish precise treatment pathways, especially for pediatric patients.”
This study was supported by the Lee Kun-Hee Pediatric Cancer and Rare Disease Research Project and the Korean National Research Foundation’s Excellent Early Career Researcher program.

[Photo] Professor Lee Sang-Yeon, Department of Otorhinolaryngology